本文将随笔者的研究进展持续更新。目前计划添加的内容有:
- 训练的数据集格式和一些技巧
- CRF的原理解释
Bonito Basecaller 概述
Bonito(中文直译为鲣鱼,ont很喜欢拿鱼的名字作为软件包的名字)是Oxford Nanopore Technology开发的一款basecaller框架(使用bonito训练的basecaller权重可以被导出至dorado,guppy等使用)。苦于互联网上对basecaller的定性描述文章较少,笔者在研究这些basecaller的架构和数据流时花费了许多的时间与精力。因此在本文中,笔者尽量以通俗易懂的语言,介绍bonito的一些数据流和数据格式等二次开发bonito(甚至是其他的纳米孔测序basecaller)需要了解的一些知识,希望能给后来者一些启发。
什么是Basecaller?
简单来讲,Basecaller是一个神经网络,他的输入是纳米孔测序时,碱基通过纳米孔时得到的电流,输出是ATCG碱基以及一些其他的调试信息。在ONT的商业套装中,电流一般被存储为fast5格式(基于hdf5)和近两年推出的pod5格式。不管是哪种格式,存储的数据结构都至少包括了以下的一些信息:
| 存储的项目 | 说明 |
|---|---|
| read_id | uuid,用于标识唯一序列用 |
| 原始电流(raw) | 从放大器中直接输出的采样值,没有单位 |
| scale,offset | 放大系数和偏移系数,通过pA_val = scale * (raw + offset)可以得到以pA为单位的电流 |
| metadata | 和测序相关的一些信息,如时间,使用的试剂盒,测序flowcell等 |
有读者可能会注意到,纳米孔测序的电流与语音识别的音频信号有些相似:比如,录入麦克风的声音也是以一维数组存储的;同时,Basecaller输出的ATCG也和语音识别网络(ASR)得到的文字转写(transcript)类似。因此,我们也可以认为纳米孔测序中的Basecaller是一个特殊的ASR网络。
Bonito的架构
以下是torchinfo输出的模型架构图([email protected], state_len=4),模型的输入为[64,5000](batchsize, current_length)
==========================================================================================
Layer (type:depth-idx) Output Shape Param #
==========================================================================================
Model [64, 1000, 1024] --
├─Sequential: 1-1 [64, 1000, 1024] --
│ └─Convolution: 2-1 [64, 4, 5000] --
│ │ └─Conv1d: 3-1 [64, 4, 5000] 24
│ │ └─Swish: 3-2 [64, 4, 5000] --
│ └─Convolution: 2-2 [64, 16, 5000] --
│ │ └─Conv1d: 3-3 [64, 16, 5000] 336
│ │ └─Swish: 3-4 [64, 16, 5000] --
│ └─Convolution: 2-3 [64, 384, 1000] --
│ │ └─Conv1d: 3-5 [64, 384, 1000] 117,120
│ │ └─Swish: 3-6 [64, 384, 1000] --
│ └─Permute: 2-4 [1000, 64, 384] --
│ └─LSTM: 2-5 [1000, 64, 384] --
│ │ └─LSTM: 3-7 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-6 [1000, 64, 384] --
│ │ └─LSTM: 3-8 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-7 [1000, 64, 384] --
│ │ └─LSTM: 3-9 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-8 [1000, 64, 384] --
│ │ └─LSTM: 3-10 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-9 [1000, 64, 384] --
│ │ └─LSTM: 3-11 [1000, 64, 384] 1,182,720
│ └─Permute: 2-10 [64, 1000, 384] --
│ └─LinearCRFEncoder: 2-11 [64, 1000, 1024] --
│ │ └─Linear: 3-12 [64, 1000, 1024] 394,240
│ │ └─Tanh: 3-13 [64, 1000, 1024] --
==========================================================================================
Total params: 6,425,320
Trainable params: 6,417,640
Non-trainable params: 7,680
Total mult-adds (G): 386.11
==========================================================================================
Input size (MB): 0.64
Forward/backward pass size (MB): 877.57
Params size (MB): 12.85
Estimated Total Size (MB): 891.06
==========================================================================================
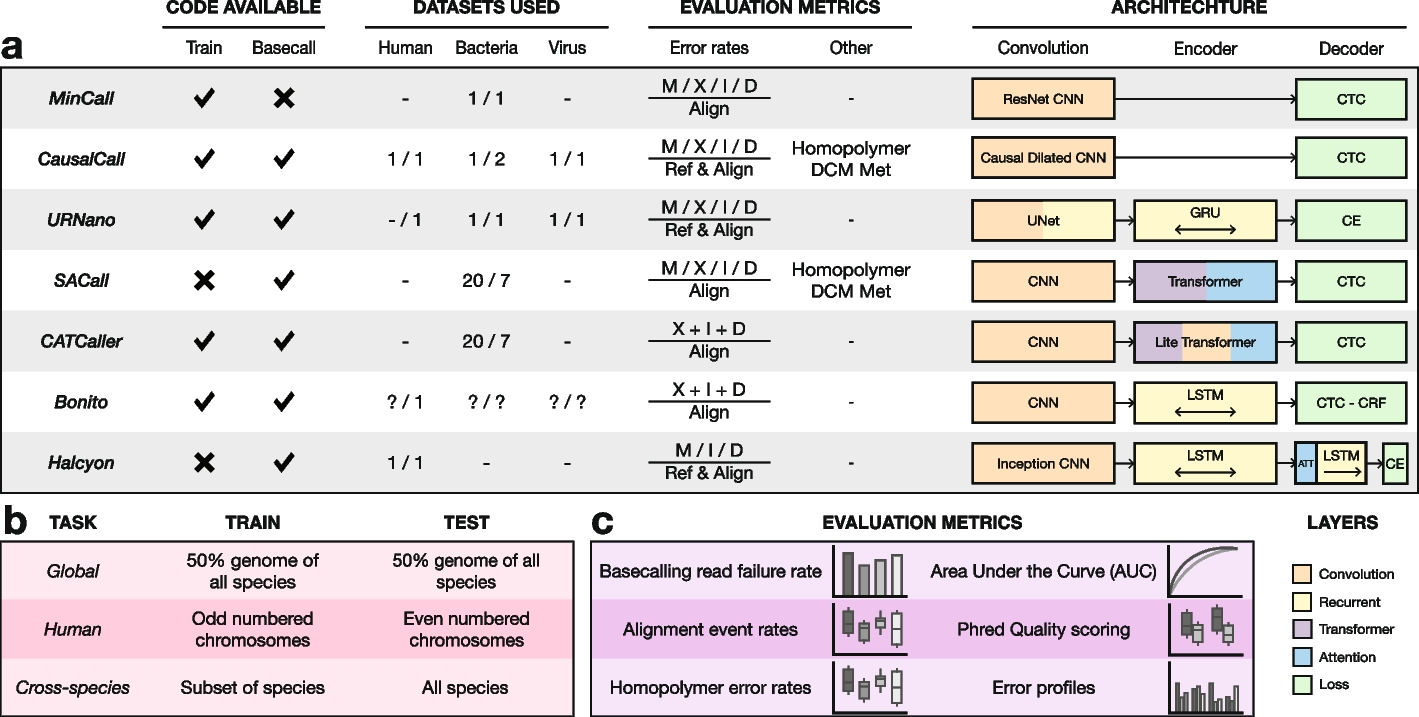
如Marcus等人文章中的图1所示,bonito(和一些常见的basecaler)的模型结构主要分为三大部份:CNN,LSTM/RNN等Encoder和CTC/CRF等Decoder。
def rnn_encoder(n_base, state_len, insize=1, first_conv_size=4, stride=5, winlen=19, activation='swish', rnn_type='lstm', features=768, scale=5.0, blank_score=None, expand_blanks=True, num_layers=5, norm=None):
rnn = layers[rnn_type]
return Serial([
conv(insize, first_conv_size, ks=5, bias=True, activation=activation, norm=norm),
conv(first_conv_size, 16, ks=5, bias=True, activation=activation, norm=norm),
conv(16, features, ks=winlen, stride=stride, bias=True, activation=activation, norm=norm),
Permute([2, 0, 1]),
*(rnn(features, features, reverse=(num_layers - i) % 2) for i in range(num_layers)),
LinearCRFEncoder(
features, n_base, state_len, activation='tanh', scale=scale,
blank_score=blank_score, expand_blanks=expand_blanks
)
])
配置文件(a.k.a config.toml)
以[email protected] (基于 bonito/bonito/models/configs/[email protected] at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub )为例:
[global_norm]
# State Length of CRF, determine how many state CRF decoder need to consider
state_len = 4
[input]
features = 1
[labels]
labels = [ "N", "A", "C", "G", "T",]
# labels of bases, N means empty state.
[qscore]
scale = 0.9356
bias = -0.1721
# bias factor of Q Score
[model]
package = "bonito.crf"
# use CRF or CTC at decoder (since bonito v0.4, only CRF is valid)
[encoder]
scale = 5.0
rnn_type = "lstm"
winlen = 19
features = 384
activation = "swish"
stride = 5
# downsample stride
blank_score = 2.0
[basecaller]
# config when basecalling, don't affect training
batchsize = 512
chunksize = 10000
overlap = 500
CNN
电流的一维数组输入后,首先会经过三次卷积以进行特征提取。卷集的实现代码位于此处,可以看到是标准的pytorch卷积实现(Conv1d — PyTorch 2.4 documentation),没有太特殊的地方。但需要注意的是,bonito为了加快运算速度,在第三层卷积设置了stride为5,令信号被下采样了5倍(5000 -> 1000)。最终,卷积层的输出为[64, 384, 1000] (batch, channel, signal_length)
bonito/bonito/nn.py at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub
@register
class Convolution(Module):
def __init__(self, insize, size, winlen, stride=1, padding=0, bias=True, activation=None, norm=None):
super().__init__()
self.conv = torch.nn.Conv1d(insize, size, winlen, stride=stride, padding=padding, bias=bias)
self.activation = layers.get(activation, lambda: activation)()
if isinstance(norm, dict):
self.norm = from_dict(norm)
elif isinstance(norm, str):
self.norm = layers[norm](size)
else:
self.norm = norm
def forward(self, x):
h = self.conv(x)
if self.norm is not None:
h = self.norm(h)
if self.activation is not None:
h = self.activation(h)
return h
def to_dict(self, include_weights=False):
res = {
"insize": self.conv.in_channels,
"size": self.conv.out_channels,
"bias": self.conv.bias is not None,
"winlen": self.conv.kernel_size[0],
"stride": self.conv.stride[0],
"padding": self.conv.padding[0],
}
if self.activation is not None:
res["activation"] = self.activation.name
if self.norm is not None:
res["norm"] = to_dict(self.norm, include_weights)
#simplify default case e.g. norm="batchnorm"
if not include_weights and self.norm.name in layers:
if res["norm"] == to_dict(layers[self.norm.name](res["size"])):
res["norm"] = self.norm.name
if include_weights:
res['params'] = {
'W': self.conv.weight, 'b': self.conv.bias if self.conv.bias is not None else []
}
return res
LSTM
经过特征提取后的信号经过一个全连接层,随后会进入LSTM层,以学习信号特征在时间上的关系。这里的LSTM也是基于标准的LSTM — PyTorch 2.4 documentation:
bonito/bonito/nn.py at 91fb1408398fb3d8188621f1486281a2baa76318 · nanoporetech/bonito · GitHub
@register
class LSTM(RNNWrapper):
def __init__(self, size, insize, bias=True, reverse=False):
super().__init__(torch.nn.LSTM, size, insize, bias=bias, reverse=reverse)
def to_dict(self, include_weights=False):
res = {
'size': self.rnn.hidden_size,
'insize': self.rnn.input_size,
'bias': self.rnn.bias,
'reverse': self.reverse,
}
if include_weights:
res['params'] = {
'iW': self.rnn.weight_ih_l0.reshape(4, self.rnn.hidden_size, self.rnn.input_size),
'sW': self.rnn.weight_hh_l0.reshape(4, self.rnn.hidden_size, self.rnn.hidden_size),
'b': self.rnn.bias_ih_l0.reshape(4, self.rnn.hidden_size)
}
return res
LSTM输出层的形状为[1000, 64, 384](signal_length, batch, channel)
CRF Encoder
数据在离开LSTM后,会进入一个全连接层,以输出一个用于CRF解码的矩阵。从代码可以看到这个Encoder做的只是把数据进行了一次非线性变换,并且对输出进行了重排。
bonito/bonito/nn.py at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub
@register
class LinearCRFEncoder(Module):
def __init__(self, insize, n_base, state_len, bias=True, scale=None, activation=None, blank_score=None, expand_blanks=True, permute=None):
super().__init__()
self.scale = scale
self.n_base = n_base
self.state_len = state_len
self.blank_score = blank_score
self.expand_blanks = expand_blanks
size = (n_base + 1) * n_base**state_len if blank_score is None else n_base**(state_len + 1)
self.linear = torch.nn.Linear(insize, size, bias=bias)
self.activation = layers.get(activation, lambda: activation)()
self.permute = permute
def forward(self, x):
if self.permute is not None:
x = x.permute(*self.permute)
scores = self.linear(x)
if self.activation is not None:
scores = self.activation(scores)
if self.scale is not None:
scores = scores * self.scale
if self.blank_score is not None and self.expand_blanks:
T, N, C = scores.shape
scores = torch.nn.functional.pad(
scores.view(T, N, C // self.n_base, self.n_base),
(1, 0, 0, 0, 0, 0, 0, 0),
value=self.blank_score
).view(T, N, -1)
return scores
整个神经网络带权重的部分到此结束。后面的CRF解码器没有包括任何权重。(因此在训练和推理时,bonito使用了不一样的解码器,后面详细解释)
CRF Decoder
CRF Decoder的核心都位于这个函数中:
def compute_scores(model, batch, beam_width=32, beam_cut=100.0, scale=1.0, offset=0.0, blank_score=2.0, reverse=False):
"""
Compute scores for model.
"""
with torch.inference_mode():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model(batch.to(dtype).to(device))
if reverse:
scores = model.seqdist.reverse_complement(scores)
with torch.cuda.device(scores.device):
sequence, qstring, moves = beam_search(
scores, beam_width=beam_width, beam_cut=beam_cut,
scale=scale, offset=offset, blank_score=blank_score
)
return {
'moves': moves,
'qstring': qstring,
'sequence': sequence,
}
在compute_scores中,scores就是前述神经网络推理出的矩阵,size为[64, 1000, 1024](batch, current_length, state),$1024=4^5(4base^{4state+1})$。接着,如果测的是RNA,则将score反过来(这也是为什么RNA训练的时候需要把reference fasta反转)。最后,调用koi(一个ONT开发的,不开源的CRF解码包,下面会详细介绍)的beam_search得到moves(输出第几个采样点解码出了碱基,用于碱基序列和电流的对其参考),qstring(Q score string,以数字编码)和sequence(序列,以数字编码)。
不开源的koi包和其开源替代
自bonito引入CRF解码开始,bonito就将beam search的函数封装进了一个不开源的ont-koi包,这导致了我们无法了解具体的CRF解码实现。但万幸的是,ont在GitHub - davidcpage/seqdist和GitHub - nanoporetech/fast-ctc-decode: Blitzing Fast CTC Beam Search Decoder这两个repo里包含了一些CRF解码的逻辑,同时,ont还在老版bonito中负责处理duplex的部分用前述的开源代码搭出了一个可用的compute_score的工作流:
def compute_scores(model, batch, reverse=False):
with torch.no_grad():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model.encoder(batch.to(dtype).to(device))
if reverse: scores = model.seqdist.reverse_complement(scores)
betas = model.seqdist.backward_scores(scores.to(torch.float32))
trans, init = model.seqdist.compute_transition_probs(scores, betas)
return {
'trans': trans.to(dtype).transpose(0, 1),
'init': init.to(dtype).unsqueeze(1),
}
def beam_search_duplex(seq1, path1, t1, b1, seq2, path2, t2, b2, alphabet='NACGT', beamsize=5, pad=40, T=0.01):
env = build_envelope(t1.shape[0], seq1, path1, t2.shape[0], seq2, path2, padding=pad)
return crf_beam_search_duplex(
t1, b1, t2, b2,
alphabet=alphabet,
beam_size=beamsize,
beam_cut_threshold=T,
envelope=env,
)
笔者同时也参考了Marcus对bonito crf解码的研究,得到了一个利用ont的开源代码实现的compute_score函数。函数主要需要修改两处,列举如下:
- 修改
backward_scores函数为开源实现(可以用bonito 0.5之前的backward_score实现):
import seqdist.sparse
from seqdist.ctc_simple import logZ_cupy, viterbi_alignments
from seqdist.core import SequenceDist, Max, Log, semiring
def backward_scores(self, scores, S: semiring=Log):
T, N, _ = scores.shape
Ms = scores.reshape(T, N, -1, self.n_base + 1)
beta_T = Ms.new_full((N, self.n_base**(self.state_len)), S.one)
return seqdist.sparse.bwd_scores_cupy(Ms, self.idx, beta_T, S, K=1)
- 修改
compute_scores函数,记得引入对应的包:
from fast_ctc_decode import crf_greedy_search
def compute_scores(model, batch, beam_width=32, beam_cut=100.0, scale=1.0, offset=0.0, blank_score=2.0, reverse=False):
"""
Compute scores for model.
"""
with torch.inference_mode():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model(batch.to(dtype).to(device))
if reverse:
scores = model.seqdist.reverse_complement(scores)
# switch dim 1 & 2
scores_pad = scores.permute(1, 0, 2)
# pad score
n_base = model.seqdist.n_base
T, N, C = scores_pad.shape
scores_pad = torch.nn.functional.pad(
scores_pad.view(T, N, C // n_base, n_base),
(1, 0, 0, 0, 0, 0, 0, 0),
value=blank_score
).view(T, N, -1)
betas = model.seqdist.backward_scores(scores_pad.to(torch.float32))
trans, init = model.seqdist.compute_transition_probs(scores_pad, betas)
trans = trans.to(torch.float32).transpose(0, 1)
init = init.to(torch.float32).unsqueeze(1)
# offload
tracebacks = trans.cpu()
init = init.cpu()
seq_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
qstring_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
moves_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
for batch_idx in range(N):
tracebacks_batch = tracebacks[batch_idx].numpy() # (T, 256, 5)
init_batch = init[batch_idx][0].numpy() # (256,)
# greedy decode, cef_beam_search dont output qstring
seq_batch, path_batch = crf_greedy_search(
network_output=tracebacks_batch,
init_state=init_batch,
alphabet="NACGT",
qstring=True,
qscale=1,
qbias=1
)
# re-encode
seq_batch_str = seq_batch[:len(seq_batch) // 2]
qstring_batch_str = seq_batch[len(seq_batch) // 2:]
seq_as_numbers = np.frombuffer(seq_batch_str.encode(), dtype=np.uint8).copy()
qstring_as_numbers = np.frombuffer(qstring_batch_str.encode(), dtype=np.uint8).copy()
seq_tensor[batch_idx, path_batch] = torch.from_numpy(seq_as_numbers[:len(path_batch)])
qstring_tensor[batch_idx, path_batch] = torch.from_numpy(qstring_as_numbers[:len(path_batch)])
moves_tensor[batch_idx, path_batch] = 1
return {
'qstring': qstring_tensor,
'sequence': seq_tensor,
'moves': moves_tensor,
}
beam_search换成了crf_greedy_search,因此basecaller的准确率可能略有下降,但根据笔者的测试,准确率仅下降约0.3%,笔者认为不完美,但可接受。此时,得到的qstring,sequence是一个长current_length的矩阵。其中部份index为0(说明这个位置并没有解码出新的碱基),剩下的index则是数字(数字编码的ATCG碱基,或q string分数)。
拼接basecaller结果和输出字符
由于神经网络的窗口大小有限,在遇到长序列的电流时,电流会按照网络配置中给定的chunksize和overlap拆成一段段的短序列。因此,在完成解码后,得到的qstring,sequence和moves需要经过stitch_results函数重新根据read_id拼接在一起。
在拼接完后,序列则会经过如path_to_str这样的解码函数被解码回碱基序列/Q Score。最后,经过格式化就可以输出为fastq,或过一次mappy之后即可输出aligned sam/bam文件了。
训练Bonito
数据集结构
参考bonito 0.8.1的官方文档,我们至少需要三个.npy文件才能构成一个训练basecaller的数据集(假设训练时用的每段电流长度为chunksize)。参考bonito的download.py,目前bonito提供了三个训练数据集,列举如下:
每个数据集下载下来并解压都包含三个.npy文件:
| Filename | Shape | 用途 |
|---|---|---|
| references.npy | (data_length, max_len_of_reference) uint8 | 存储每个信号chunk对应的序列,用诸如{'A': 1, 'C': 2, 'G': 3, 'T': 4}的规则编码ATCG,余下位置用0填充 |
| reference_lengths.npy | (data_length,) uint8 | 存储每个信号chunk对应的序列的长度(不pad0的部分) |
| chunks.npy | (data_length, chunksize) float32 | 存储每个信号chunk的电流讯号 |
可选的,数据集文件夹中也可以包含一个名为validation_sets的文件夹,如果存在此文件夹,里面的references.npy,reference_lengths.npy,chunks.npy将作为验证集使用。
训练相关的命令
# bonito train
positional arguments:
training_directory
optional arguments:
-h, --help show this help message and exit
--config CONFIG
--pretrained PRETRAINED
--directory DIRECTORY
--device DEVICE
--lr LR
--seed SEED
--epochs EPOCHS
--batch BATCH
--chunks CHUNKS
--valid-chunks VALID_CHUNKS
--no-amp
-f, --force
--restore-optim
--nondeterministic
--save-optim-every SAVE_OPTIM_EVERY
--grad-accum-split GRAD_ACCUM_SPLIT
--quantile-grad-clip
--num-workers NUM_WORKERS
训练时的loss函数计算
和推理时使用的CRF不同,bonito在训练时,使用的解码器和loss函数并不是先前提到的compute_score函数。
def decode_batch(self, x):
scores = self.seqdist.posteriors(x.to(torch.float32)) + 1e-8
tracebacks = self.seqdist.viterbi(scores.log()).to(torch.int16).T
return [self.seqdist.path_to_str(x) for x in tracebacks.cpu().numpy()]
而是用seqdist的viterbi search直接得出了序列,然后跟标准参考序列比较得出了一个准确率。
训练命令
bonito train \
--config config.toml \ # 前面提到的模型config,chunksize等会从数据集里自动读取
--device cuda:0 \ # 显卡
--epochs 5 # epoch
--lr 5e-4 \ # 学习率
--batch 96 \ # batchsize
--pretrained [email protected] \ # 现有模型,可以从ont提供的模型选,或者传入其他模型所在的目录
--directory dataset_dir/ \ # chunks.npy等所在的文件夹
model_dir/
