This article will be continuously updated as my research progresses. The currently planned additions include:
- Training dataset format and some techniques
- Explanation of the principles of CRF
Overview of Bonito Basecaller
Bonito, named after the bonito fish, is a basecaller framework developed by Oxford Nanopore Technology. The trained basecaller weights can be exported to dorado, guppy, etc. Due to the scarcity of qualitative descriptions of basecallers on the internet, I have spent a lot of time and effort studying the architecture and data flow of these basecallers. Therefore, in this article, I will try to explain, in an easy-to-understand way, the data flow and data formats of Bonito, as well as some knowledge needed for the development of Bonito (or even other nanopore sequencing basecallers). I hope to provide some inspiration for those who come after.
What is a Basecaller?
To make it simple, a “basecaller” is a neural network. Its input is the current signal obtained from nanopore translocation, and its output is the ATCG bases and some other debugging information (in fasta, fastq or bam format). In ONT’s devices, the current is generally stored in fast5 format (based on hdf5) and the recently released pod5 format. Regardless of the format, the stored data structure includes at least the following information:
| Stored Item | Description |
|---|---|
| read_id | uuid, used to uniquely identify the sequence |
| raw current | Sampled values directly output from the amplifier, without units |
| scale, offset | Amplification and offset coefficients, the current in pA can be obtained by pA_val = scale * (raw + offset) |
| metadata | Some information related to sequencing, such as time, the kit used, the sequencing flowcell, etc. |
Some readers may notice that the current signal from nanopore sequencing is somewhat similar to the audio signal in speech recognition. For example, the sound recorded by a microphone is also stored as a one-dimensional array. At the same time, the ATCG output by the basecaller is also similar to the transcript obtained by the speech recognition network (ASR). Therefore, we can also consider the basecaller in nanopore sequencing as a special ASR network.
Architecture of Bonito
The following is the model architecture diagram output by torchinfo ([email protected], state_len=4), the input of the model is [64,5000] (batchsize, current_length)
==========================================================================================
Layer (type:depth-idx) Output Shape Param #
==========================================================================================
Model [64, 1000, 1024] --
├─Sequential: 1-1 [64, 1000, 1024] --
│ └─Convolution: 2-1 [64, 4, 5000] --
│ │ └─Conv1d: 3-1 [64, 4, 5000] 24
│ │ └─Swish: 3-2 [64, 4, 5000] --
│ └─Convolution: 2-2 [64, 16, 5000] --
│ │ └─Conv1d: 3-3 [64, 16, 5000] 336
│ │ └─Swish: 3-4 [64, 16, 5000] --
│ └─Convolution: 2-3 [64, 384, 1000] --
│ │ └─Conv1d: 3-5 [64, 384, 1000] 117,120
│ │ └─Swish: 3-6 [64, 384, 1000] --
│ └─Permute: 2-4 [1000, 64, 384] --
│ └─LSTM: 2-5 [1000, 64, 384] --
│ │ └─LSTM: 3-7 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-6 [1000, 64, 384] --
│ │ └─LSTM: 3-8 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-7 [1000, 64, 384] --
│ │ └─LSTM: 3-9 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-8 [1000, 64, 384] --
│ │ └─LSTM: 3-10 [1000, 64, 384] 1,182,720
│ └─LSTM: 2-9 [1000, 64, 384] --
│ │ └─LSTM: 3-11 [1000, 64, 384] 1,182,720
│ └─Permute: 2-10 [64, 1000, 384] --
│ └─LinearCRFEncoder: 2-11 [64, 1000, 1024] --
│ │ └─Linear: 3-12 [64, 1000, 1024] 394,240
│ │ └─Tanh: 3-13 [64, 1000, 1024] --
==========================================================================================
Total params: 6,425,320
Trainable params: 6,417,640
Non-trainable params: 7,680
Total mult-adds (G): 386.11
==========================================================================================
Input size (MB): 0.64
Forward/backward pass size (MB): 877.57
Params size (MB): 12.85
Estimated Total Size (MB): 891.06
==========================================================================================
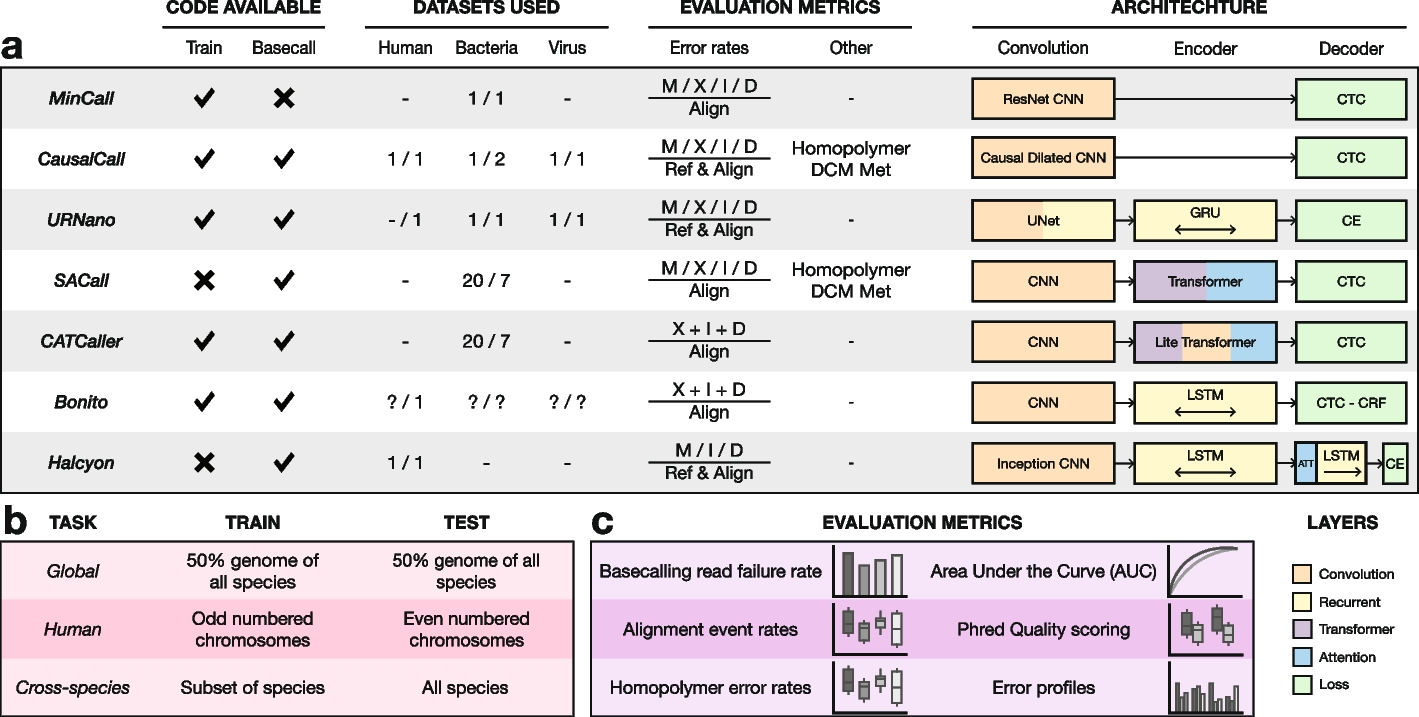
As shown in Figure 1 of Marcus et al.’s article, the model structure of Bonito (and some common basecallers) is mainly divided into three parts: CNN, LSTM/RNN, etc. Encoder, and CTC/CRF, etc. Decoder.
def rnn_encoder(n_base, state_len, insize=1, first_conv_size=4, stride=5, winlen=19, activation='swish', rnn_type='lstm', features=768, scale=5.0, blank_score=None, expand_blanks=True, num_layers=5, norm=None):
rnn = layers[rnn_type]
return Serial([
conv(insize, first_conv_size, ks=5, bias=True, activation=activation, norm=norm),
conv(first_conv_size, 16, ks=5, bias=True, activation=activation, norm=norm),
conv(16, features, ks=winlen, stride=stride, bias=True, activation=activation, norm=norm),
Permute([2, 0, 1]),
*(rnn(features, features, reverse=(num_layers - i) % 2) for i in range(num_layers)),
LinearCRFEncoder(
features, n_base, state_len, activation='tanh', scale=scale,
blank_score=blank_score, expand_blanks=expand_blanks
)
])
Configuration File (a.k.a config.toml)
Take [email protected] (based on bonito/bonito/models/configs/[email protected] at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub ) as an example:
[global_norm]
# State Length of CRF, determine how many state CRF decoder need to consider
state_len = 4
[input]
features = 1
[labels]
labels = [ "N", "A", "C", "G", "T",]
# labels of bases, N means empty state.
[qscore]
scale = 0.9356
bias = -0.1721
# bias factor of Q Score
[model]
package = "bonito.crf"
# use CRF or CTC at decoder (since bonito v0.4, only CRF is valid)
[encoder]
scale = 5.0
rnn_type = "lstm"
winlen = 19
features = 384
activation = "swish"
stride = 5
# downsample stride
blank_score = 2.0
[basecaller]
# config when basecalling, don't affect training
batchsize = 512
chunksize = 10000
overlap = 500
CNN
After the one-dimensional array of current is input, it will first go through three convolutions for feature extraction. The implementation code of the convolution is located here, it can be seen that it is a standard pytorch convolution implementation (Conv1d — PyTorch 2.4 documentation), there is nothing too special. But it should be noted that in order to speed up the calculation, Bonito sets the stride to 5 in the third convolutional layer, which downsamples the signal by 5 times (5000 -> 1000). Finally, the output of the convolutional layer is [64, 384, 1000] (batch, channel, signal_length).
bonito/bonito/nn.py at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub
@register
class Convolution(Module):
def __init__(self, insize, size, winlen, stride=1, padding=0, bias=True, activation=None, norm=None):
super().__init__()
self.conv = torch.nn.Conv1d(insize, size, winlen, stride=stride, padding=padding, bias=bias)
self.activation = layers.get(activation, lambda: activation)()
if isinstance(norm, dict):
self.norm = from_dict(norm)
elif isinstance(norm, str):
self.norm = layers[norm](size)
else:
self.norm = norm
def forward(self, x):
h = self.conv(x)
if self.norm is not None:
h = self.norm(h)
if self.activation is not None:
h = self.activation(h)
return h
def to_dict(self, include_weights=False):
res = {
"insize": self.conv.in_channels,
"size": self.conv.out_channels,
"bias": self.conv.bias is not None,
"winlen": self.conv.kernel_size[0],
"stride": self.conv.stride[0],
"padding": self.conv.padding[0],
}
if self.activation is not None:
res["activation"] = self.activation.name
if self.norm is not None:
res["norm"] = to_dict(self.norm, include_weights)
#simplify default case e.g. norm="batchnorm"
if not include_weights and self.norm.name in layers:
if res["norm"] == to_dict(layers[self.norm.name](res["size"])):
res["norm"] = self.norm.name
if include_weights:
res['params'] = {
'W': self.conv.weight, 'b': self.conv.bias if self.conv.bias is not None else []
}
return res
LSTM
After feature extraction, the signal passes through a fully connected layer, and then enters the LSTM layer to learn the temporal relationship of signal features. The LSTM here is also based on the standard LSTM — PyTorch 2.4 documentation:
bonito/bonito/nn.py at 91fb1408398fb3d8188621f1486281a2baa76318 · nanoporetech/bonito · GitHub
@register
class LSTM(RNNWrapper):
def __init__(self, size, insize, bias=True, reverse=False):
super().__init__(torch.nn.LSTM, size, insize, bias=bias, reverse=reverse)
def to_dict(self, include_weights=False):
res = {
'size': self.rnn.hidden_size,
'insize': self.rnn.input_size,
'bias': self.rnn.bias,
'reverse': self.reverse,
}
if include_weights:
res['params'] = {
'iW': self.rnn.weight_ih_l0.reshape(4, self.rnn.hidden_size, self.rnn.input_size),
'sW': self.rnn.weight_hh_l0.reshape(4, self.rnn.hidden_size, self.rnn.hidden_size),
'b': self.rnn.bias_ih_l0.reshape(4, self.rnn.hidden_size)
}
return res
The shape of the LSTM output layer is [1000, 64, 384] (signal_length, batch, channel)
CRF Encoder
After the data leaves the LSTM, it will enter a fully connected layer to output a matrix for CRF decoding. From the code, we can see that this encoder only performs a non-linear transformation on the data, and rearranges the output.
bonito/bonito/nn.py at 0c7fcceeeca16e300ba427d737282b33d3cb8ec9 · nanoporetech/bonito · GitHub
@register
class LinearCRFEncoder(Module):
def __init__(self, insize, n_base, state_len, bias=True, scale=None, activation=None, blank_score=None, expand_blanks=True, permute=None):
super().__init__()
self.scale = scale
self.n_base = n_base
self.state_len = state_len
self.blank_score = blank_score
self.expand_blanks = expand_blanks
size = (n_base + 1) * n_base**state_len if blank_score is None else n_base**(state_len + 1)
self.linear = torch.nn.Linear(insize, size, bias=bias)
self.activation = layers.get(activation, lambda: activation)()
self.permute = permute
def forward(self, x):
if self.permute is not None:
x = x.permute(*self.permute)
scores = self.linear(x)
if self.activation is not None:
scores = self.activation(scores)
if self.scale is not None:
scores = scores * self.scale
if self.blank_score is not None and self.expand_blanks:
T, N, C = scores.shape
scores = torch.nn.functional.pad(
scores.view(T, N, C // self.n_base, self.n_base),
(1, 0, 0, 0, 0, 0, 0, 0),
value=self.blank_score
).view(T, N, -1)
return scores
The part with weights in the entire neural network ends here. The following CRF decoder does not include any weights. (Therefore, during training and inference, Bonito uses different decoders, which will be explained in detail later)
CRF Decoder
The core of the CRF Decoder is in this function:
def compute_scores(model, batch, beam_width=32, beam_cut=100.0, scale=1.0, offset=0.0, blank_score=2.0, reverse=False):
"""
Compute scores for model.
"""
with torch.inference_mode():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model(batch.to(dtype).to(device))
if reverse:
scores = model.seqdist.reverse_complement(scores)
with torch.cuda.device(scores.device):
sequence, qstring, moves = beam_search(
scores, beam_width=beam_width, beam_cut=beam_cut,
scale=scale, offset=offset, blank_score=blank_score
)
return {
'moves': moves,
'qstring': qstring,
'sequence': sequence,
}
In compute_scores, scores is the matrix inferred by the aforementioned neural network, with a size of [64, 1000, 1024] (batch, current_length, state), $1024=4^5(4base^{4state+1})$. Then, if RNA is being sequenced, the score is reversed (this is also why the reference fasta needs to be reversed when training RNA). Finally, call koi (a non-open source CRF decoding package developed by ONT, which will be explained in detail below) beam_search to get moves (output which sampling point decoded a base, used for base sequence and current alignment reference), qstring (Q score string, encoded in numbers) and sequence (sequence, encoded in numbers).
Close-sourced koi Package and Its Open Source Alternatives
Since Bonito introduced CRF decoding, Bonito has encapsulated the beam search function into a close-sourced ont-koi package, which makes it impossible for us to understand the specific implementation of CRF decoding. But fortunately, ONT has included some CRF decoding logic in these two repos: GitHub - davidcpage/seqdist and GitHub - nanoporetech/fast-ctc-decode: Blitzing Fast CTC Beam Search Decoder. At the same time, ONT used the aforementioned open-source code in the old version of Bonito to build a usable compute_score workflow to handle the duplex part:
def compute_scores(model, batch, reverse=False):
with torch.no_grad():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model.encoder(batch.to(dtype).to(device))
if reverse: scores = model.seqdist.reverse_complement(scores)
betas = model.seqdist.backward_scores(scores.to(torch.float32))
trans, init = model.seqdist.compute_transition_probs(scores, betas)
return {
'trans': trans.to(dtype).transpose(0, 1),
'init': init.to(dtype).unsqueeze(1),
}
def beam_search_duplex(seq1, path1, t1, b1, seq2, path2, t2, b2, alphabet='NACGT', beamsize=5, pad=40, T=0.01):
env = build_envelope(t1.shape[0], seq1, path1, t2.shape[0], seq2, path2, padding=pad)
return crf_beam_search_duplex(
t1, b1, t2, b2,
alphabet=alphabet,
beam_size=beamsize,
beam_cut_threshold=T,
envelope=env,
)
I have also referred to Marcus’s research on Bonito CRF decoding, and obtained a compute_score function implemented using ONT’s open source code. The function mainly needs to be modified in two places, as listed below:
- Modify the
backward_scoresfunction to an open source implementation (the backward_score implementation before Bonito 0.5 can be used):
import seqdist.sparse
from seqdist.ctc_simple import logZ_cupy, viterbi_alignments
from seqdist.core import SequenceDist, Max, Log, semiring
def backward_scores(self, scores, S: semiring=Log):
T, N, _ = scores.shape
Ms = scores.reshape(T, N, -1, self.n_base + 1)
beta_T = Ms.new_full((N, self.n_base**(self.state_len)), S.one)
return seqdist.sparse.bwd_scores_cupy(Ms, self.idx, beta_T, S, K=1)
- Modify the
compute_scoresfunction, remember to import the corresponding packages:
from fast_ctc_decode import crf_greedy_search
def compute_scores(model, batch, beam_width=32, beam_cut=100.0, scale=1.0, offset=0.0, blank_score=2.0, reverse=False):
"""
Compute scores for model.
"""
with torch.inference_mode():
device = next(model.parameters()).device
dtype = torch.float16 if half_supported() else torch.float32
scores = model(batch.to(dtype).to(device))
if reverse:
scores = model.seqdist.reverse_complement(scores)
# switch dim 1 & 2
scores_pad = scores.permute(1, 0, 2)
# pad score
n_base = model.seqdist.n_base
T, N, C = scores_pad.shape
scores_pad = torch.nn.functional.pad(
scores_pad.view(T, N, C // n_base, n_base),
(1, 0, 0, 0, 0, 0, 0, 0),
value=blank_score
).view(T, N, -1)
betas = model.seqdist.backward_scores(scores_pad.to(torch.float32))
trans, init = model.seqdist.compute_transition_probs(scores_pad, betas)
trans = trans.to(torch.float32).transpose(0, 1)
init = init.to(torch.float32).unsqueeze(1)
# offload
tracebacks = trans.cpu()
init = init.cpu()
seq_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
qstring_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
moves_tensor = torch.zeros((N, T), dtype=torch.uint8, device='cpu')
for batch_idx in range(N):
tracebacks_batch = tracebacks[batch_idx].numpy() # (T, 256, 5)
init_batch = init[batch_idx][0].numpy() # (256,)
# greedy decode, cef_beam_search dont output qstring
seq_batch, path_batch = crf_greedy_search(
network_output=tracebacks_batch,
init_state=init_batch,
alphabet="NACGT",
qstring=True,
qscale=1,
qbias=1
)
# re-encode
seq_batch_str = seq_batch[:len(seq_batch) // 2]
qstring_batch_str = seq_batch[len(seq_batch) // 2:]
seq_as_numbers = np.frombuffer(seq_batch_str.encode(), dtype=np.uint8).copy()
qstring_as_numbers = np.frombuffer(qstring_batch_str.encode(), dtype=np.uint8).copy()
seq_tensor[batch_idx, path_batch] = torch.from_numpy(seq_as_numbers[:len(path_batch)])
qstring_tensor[batch_idx, path_batch] = torch.from_numpy(qstring_as_numbers[:len(path_batch)])
moves_tensor[batch_idx, path_batch] = 1
return {
'qstring': qstring_tensor,
'sequence': seq_tensor,
'moves': moves_tensor,
}
beam_search with crf_greedy_search. Therefore, the accuracy of the basecaller may be slightly reduced. However, according to my tests, the accuracy only decreases by about 0.3%, which I think is not perfect but acceptable.At this point, the obtained qstring, sequence is a matrix with a length of current_length. Some indices are 0 (indicating that no new bases were decoded at this position), and the remaining indices are numbers (number-encoded ATCG bases or q string scores).
Stitching Basecaller Results and Outputting Characters
Due to the limited window size of the neural network, when encountering long sequences of current, the current will be split into short sequences according to the chunksize and overlap given in the network configuration. Therefore, after completing the decoding, the obtained qstring, sequence and moves need to be re-stitched together according to read_id by the stitch_results function.
After stitching, the sequence will be decoded back to the base sequence/Q Score through decoding functions such as path_to_str. Finally, after formatting, it can be output as fastq, or after going through mappy once, it can be output as an aligned sam/bam file.
Training Bonito
Dataset Structure
Referencing the official documentation of Bonito 0.8.1, we need at least three .npy files to construct a training basecaller dataset (assuming that the length of each current segment used during training is chunksize). Referencing Bonito’s download.py, Bonito currently provides three training datasets, listed below:
Each downloaded and decompressed dataset contains three .npy files:
| Filename | Shape | Purpose |
|---|---|---|
| references.npy | (data_length, max_len_of_reference) uint8 | Stores the sequence corresponding to each signal chunk, encoded using rules such as {'A': 1, 'C': 2, 'G': 3, 'T': 4} for ATCG, with the remaining positions padded with 0 |
| reference_lengths.npy | (data_length,) uint8 | Stores the length of the sequence corresponding to each signal chunk (the part not padded with 0) |
| chunks.npy | (data_length, chunksize) float32 | Stores the current signal of each signal chunk |
Optionally, the dataset folder can also contain a folder named validation_sets. If this folder exists, the references.npy, reference_lengths.npy, and chunks.npy inside it will be used as the validation set.
Training-related Commands
# bonito train
positional arguments:
training_directory
optional arguments:
-h, --help show this help message and exit
--config CONFIG
--pretrained PRETRAINED
--directory DIRECTORY
--device DEVICE
--lr LR
--seed SEED
--epochs EPOCHS
--batch BATCH
--chunks CHUNKS
--valid-chunks VALID_CHUNKS
--no-amp
-f, --force
--restore-optim
--nondeterministic
--save-optim-every SAVE_OPTIM_EVERY
--grad-accum-split GRAD_ACCUM_SPLIT
--quantile-grad-clip
--num-workers NUM_WORKERS
Loss Function Calculation During Training
Unlike the CRF used during inference, Bonito does not use the previously mentioned compute_score function as the decoder and loss function during training.
def decode_batch(self, x):
scores = self.seqdist.posteriors(x.to(torch.float32)) + 1e-8
tracebacks = self.seqdist.viterbi(scores.log()).to(torch.int16).T
return [self.seqdist.path_to_str(x) for x in tracebacks.cpu().numpy()]
Instead, it uses seqdist’s viterbi search to directly obtain the sequence, and then compares it with the standard reference sequence to obtain an accuracy.
Training Command
bonito train \
--config config.toml \ # The model config mentioned earlier, chunksize, etc., will be automatically read from the dataset
--device cuda:0 \ # GPU
--epochs 5 # epoch
--lr 5e-4 \ # learning rate
--batch 96 \ # batchsize
--pretrained [email protected] \ # Existing model, you can select from the models provided by ONT, or pass in the directory where other models are located
--directory dataset_dir/ \ # Folder containing chunks.npy, etc.
model_dir/
This command initiates the training process. Let’s break down the key components:
bonito train: This is the main command to start the training of a Bonito model.--config config.toml: This specifies the configuration file for the model. This file defines various parameters like the network architecture, number of states for CRF, input/label configurations, learning rate, and more. Theconfig.tomlfile was described in detail earlier.--device cuda:0: This argument tells Bonito to use the first CUDA-enabled GPU (if available). If you have multiple GPUs, you can specify a different ID (e.g.,cuda:1). If you want to train on CPU, you can usecpuas an argument, but training on GPU will be significantly faster.--epochs 5: This sets the number of training epochs, indicating how many times the entire training dataset will be passed through the model.--lr 5e-4: This sets the learning rate for the optimizer. A lower learning rate can result in finer tuning of the weights but may also require more training epochs.--batch 96: This specifies the batch size, i.e., the number of training examples that the model will process in each iteration. Larger batch sizes can utilize GPU resources more efficiently, but may also lead to memory issues, particularly with smaller GPUs.--pretrained [email protected]: This crucial parameter specifies the model that will be used as a starting point. This leverages transfer learning, enabling you to train more quickly and effectively. You can use ONT’s pre-trained models, like the example provided ([email protected]) or point to a custom pre-trained model.--directory dataset_dir/: This points to the directory containing the training dataset. Bonito expects the training data to be in the format explained in the “Dataset Structure” section. It will automatically load thechunks.npy,references.npy, andreference_lengths.npyfrom the provided directory, and potentially validation data from thevalidation_setssubfolder.model_dir/: Finally,model_dir/is the directory where the trained model (including model weights) and training checkpoints will be saved.
